Testing your Ansible roles could get tedious, running some playbooks against your role over and over again, after you’ve done changes and just miss out the ONE combination of parameters, that will lead to a breaking change is both time consuming and nerve wracking. Ansible Molecule gives you some nice tooling at hand to party automate your test runs. Combining them with Github Actions as CI/CD Pipeline, will run your tests completely automated, as soon as you push to your Github repository.
If you want to follow along more easily, you may want to clone my cookbooks repository. You can find my molecule test setup there under ansible/molecule.
Prerequisites:
- Your code repository must be on Github in order to use Github Actions.
- If you have higher demands on your Github Actions in order of performance, storage space or just pricing (Github Actions is only free for x minutes runtime each month, see here), you may need to setup your self-hosted Github “runner” Host / VM. Follow this instructions here to setup such a runner.
- In order to run the molecule testing locally, you need molecule installed (this is part of this article). Also you need docker or podman installed (this is NOT part of this article, you can find instructions on installing docker here). Last but not least, you need to have Python3 + pip installed on your local environment.
Install and setup Molecule
Before we head over to Github Actions, we want to setup Molecule locally (either on your workstation with Linux / WSL or on a VM running Linux). The actual installation of Molecule can be done using pip:
sudo pip3 install "molecule[lint,docker]"This will install Molecule together with yamllint and ansible-lint. If you plan not to use Docker as your container runtime (in favour for podman for example), you can leave ,docker from the install command. After the installation, you can check the installation by asking for the version, that we have installed.
molecule --version
molecule 3.5.2 using python 3.8
ansible:2.12.1
delegated:3.5.2 from molecule
docker:1.1.0 from molecule_docker requiring collections: community.docker>=1.9.1Molecule is used for testing Ansible roles primarily, using the commandline molecule, we could initialize a complete (empty) role structure as we would normally do using ansible-galaxy. The only difference is, that molecule init role, will also create us a default scenario for a Molecule test. So let’s initialize our role issuing the following command. Again, if you plan to use a different container driver, you have to specify it following the -d parameter.
molecule init role molecule_test_role -d docker
INFO Initializing new role molecule-test-role...
No config file found; using defaults
- Role molecule-test-role was created successfully
INFO Initialized role in /cookbooks/ansible/molecule/molecule_test_role successfully.Let’s have a look what molecule just did:
cd molecule_test_role
tree
.
├── README.md
├── defaults
│ └── main.yml
├── files
├── handlers
│ └── main.yml
├── meta
│ └── main.yml
├── molecule
│ └── default
│ ├── converge.yml
│ ├── molecule.yml
│ └── verify.yml
├── tasks
│ └── main.yml
├── templates
├── tests
│ ├── inventory
│ └── test.yml
└── vars
└── main.ymlThe only difference from initializing a role with the ansible-galaxy command is the folder molecule and it’s subfolders. We can find the so called scnenarios in there. There was a default scenario created for us. Let’s have a look into the molecule.yml file, which includes the central configuration of our test scenario:
cat molecule/default/molecule.yml
---
dependency:
name: galaxy
driver:
name: docker
platforms:
- name: instance
image: docker.io/pycontribs/centos:8
pre_build_image: true
provisioner:
name: ansible
verifier:
name: ansibleYou maybe already found out by the pip installation command, that we installed ansible-lint alongside with Molecule. Yes, you can also do linting during your molecule test. You can configure Molecule which linters to use, by adding the following lines to the molecule.yml file (at the bottom). If you do linting differently, you can skip this step.
lint: |
set -e
yamllint .
ansible-lint .The first time, we’re running the molecule lint, we will get some errors regarding the .travis.yml file in our project root directory. As we don’t plan to use Travis, we can remove the file. Now let’s see how to lint:
molecule lint
...
WARNING Skipping, missing the requirements file.
WARNING Skipping, missing the requirements file.
INFO Running default > lint
COMMAND: set -e
yamllint .
ansible-lint .
...
meta-incorrect: Should change default metadata: author
meta/main.yml:1
...You will see some more errors regarding the meta/main.yml. So let’s quickly add an author and other informations to the file. You can replace the content with the following for the sake of our demo:
galaxy_info:
author: Philip Haberkern
description: Molecule testing role
company: thedatabaseme
license: MIT
min_ansible_version: 2.9
platforms:
- name: redhat
versions:
- 8
- name: ubuntu
versions:
- 20.04
galaxy_tags: []
dependencies: []
Now the linting should work without warnings or errors.
It’s time to talk about, what we actually want to achieve by using our Ansible role. In this demo, we want to install an Apache webserver (httpd) on a Redhat 8 OS. For this OS, we have to get our Docker image for. You’re able to provide several platforms per scenario in Molecule. But we only have 500 MB of Storage available in our “free” Github Action, so for the moment, we will stick with one Image. In order to get Molecule to provide the proper testing environment, we have to define our platform setup in the molecule.yml.
If you want to use systemd within the container, you need to start the container as “privileged” and add the mountpoints in the example below. The reason I leave this out is again the limitation of the storage in the “free” plan of Github Actions. The container image with enabled systemd that I’ve found, is way to big for this limitation. I’ve left the privileged configuration commented out. So if you’re free of the storage limits or you’re using a self-hosten Github runner, you can use the image of Jeff Geerling.
So let’s change the platform part of our molecule.yml like here:
driver:
name: docker
### INSERT HERE
platforms:
- name: instance
image: registry.access.redhat.com/ubi8/ubi-init
pre_build_image: true
# - name: ubuntu
# image: geerlingguy/docker-ubuntu2004-ansible
# tmpfs:
# - /run
# - /tmp
# volumes:
# - /sys/fs/cgroup:/sys/fs/cgroup:ro
# privileged: true
# capabilities:
# - SYS_ADMIN
# command: "/sbin/init"
# pre_build_image: true
### TO HERE
provisioner:
name: ansibleOk, now we make a first try if our test Docker environment will get started by triggering the command molecule create. This will download the needed Docker images and run the containers with the configuration we made.
molecule create
...
INFO Running default > create
INFO Sanity checks: 'docker'
PLAY [Create] ******************************************************************
TASK [Log into a Docker registry] **********************************************
skipping: [localhost] => (item=None)
skipping: [localhost]
TASK [Check presence of custom Dockerfiles] ************************************
ok: [localhost] => (item={'image': 'registry.access.redhat.com/ubi8/ubi-init', 'name': 'instance', 'pre_build_image': True})
TASK [Create Dockerfiles from image names] *************************************
skipping: [localhost] => (item={'image': 'registry.access.redhat.com/ubi8/ubi-init', 'name': 'instance', 'pre_build_image': True})
TASK [Discover local Docker images] ********************************************
ok: [localhost] => (item={'changed': False, 'skipped': True, 'skip_reason': 'Conditional result was False', 'item': {'image': 'registry.access.redhat.com/ubi8/ubi-init', 'name': 'instance', 'pre_build_image': True}, 'ansible_loop_var': 'item', 'i': 0, 'ansible_index_var': 'i'})
TASK [Build an Ansible compatible image (new)] *********************************
skipping: [localhost] => (item=molecule_local/registry.access.redhat.com/ubi8/ubi-init)
TASK [Create docker network(s)] ************************************************
TASK [Determine the CMD directives] ********************************************
ok: [localhost] => (item={'image': 'registry.access.redhat.com/ubi8/ubi-init', 'name': 'instance', 'pre_build_image': True})
TASK [Create molecule instance(s)] *********************************************
changed: [localhost] => (item=instance)
TASK [Wait for instance(s) creation to complete] *******************************
FAILED - RETRYING: [localhost]: Wait for instance(s) creation to complete (300 retries left).
changed: [localhost] => (item={'failed': 0, 'started': 1, 'finished': 0, 'ansible_job_id': '652042360375.26348', 'results_file': '/home/phaberke/.ansible_async/652042360375.26348', 'changed': True, 'item': {'image': 'registry.access.redhat.com/ubi8/ubi-init', 'name': 'instance', 'pre_build_image': True}, 'ansible_loop_var': 'item'})
PLAY RECAP *********************************************************************
localhost : ok=5 changed=2 unreachable=0 failed=0 skipped=4 rescued=0 ignored=0
INFO Running default > prepare
WARNING Skipping, prepare playbook not configured.After all is done, we should also be able to find our container named instance within a docker ps and a molecule list command output.
docker ps
CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES
323f975a8775 registry.access.redhat.com/ubi8/ubi-init "bash -c 'while true…" 2 minutes ago Up 2 minutes instance
molecule list
INFO Running default > list
╷ ╷ ╷ ╷ ╷
Instance Name │ Driver Name │ Provisioner Name │ Scenario Name │ Created │ Converged
╶───────────────┼─────────────┼──────────────────┼───────────────┼─────────┼───────────╴
instance │ docker │ ansible │ default │ true │ false
╵ ╵ ╵ ╵ ╵Run Molecule test
So now that we’re done with the setup of molecule, we could begin in running our test environment and the actual test.
Normally a complete test scenario will go through the following steps when you run molecule test from your projects root directory:
- lint
- destroy
- dependency
- syntax
- create
- prepare
- converge
- idempotence
- side_effect
- verify
- destroy
You can adapt these steps also within the molecule.yml of your scenario by giving a new sequence under the test_sequence parameter. If you want to prevent Molecule from destroying your container setup after a finished test, you can always add the parameter --destroy=never. This will prevent the deletion of the container, regardless if you have specified it within the molecule.yml.
Below you can find a short play, that you can add into your task/main.yml file. This represents our actual role which will install the Apache Webserver.
---
- name: Be patient
debug:
msg: "Apache Webserver will be installed now!"
- name: Installing Webserver
yum:
name: httpd
state: present
when: ansible_os_family == 'RedHat'
Now, issue a complete test scenario run using molecule test, the output should look something like this:
molecule test
...
PLAY [Converge] ****************************************************************
TASK [Gathering Facts] *********************************************************
ok: [instance]
TASK [Include molecule_test_role] **********************************************
TASK [molecule_test_role : Be patient] *****************************************
ok: [instance] => {
"msg": "Apache Webserver will be installed now!"
}
TASK [molecule_test_role : Installing Webserver] *******************************
changed: [instance]
PLAY RECAP *********************************************************************
instance : ok=3 changed=1 unreachable=0 failed=0 skipped=0 rescued=0 ignored=0
INFO Running default > idempotence
...Create Github Action workflow
It’s time to get this finished right? So let’s have a look to the action we want to use within our workflow. You can have a look here if you want to get more details on this particular Molecule action.
To get Github to run an action on a preconfigured condition (a push to the repository for instance), you need to create a YAML file under the .github folder in your projects root directory. I’ve named mine molecule.yml. Add the following content to this file, commit and push it.
name: Molecule
on:
push:
branches:
- master
pull_request:
branches:
- master
jobs:
molecule:
runs-on: ubuntu-latest
steps:
- name: Checkout
uses: actions/checkout@v2
with:
path: "${{ github.repository }}"
- name: Molecule
uses: gofrolist/molecule-action@v2
with:
molecule_command: test
molecule_args: -d docker
molecule_working_dir: thedatabaseme/cookbooks/ansible/molecule/molecule_test_role
env:
ANSIBLE_FORCE_COLOR: '1'
Caution: You need to adjust the parameter molecule_working_dir to your needs. First, this parameter is only needed, if your role root directory does not equal your repository root directory. If so, you need to adjust your repositories owner / username, the repository name and the path to your role accordingly. It should look then something like this: <REPO_USER>/<REPO_NAME>/<PATH>/<TO>/<ROLE>.
Now head over to your Github repository page and select the Action tab there. You should see a workflow named Molecule already running. Click on it for more details.

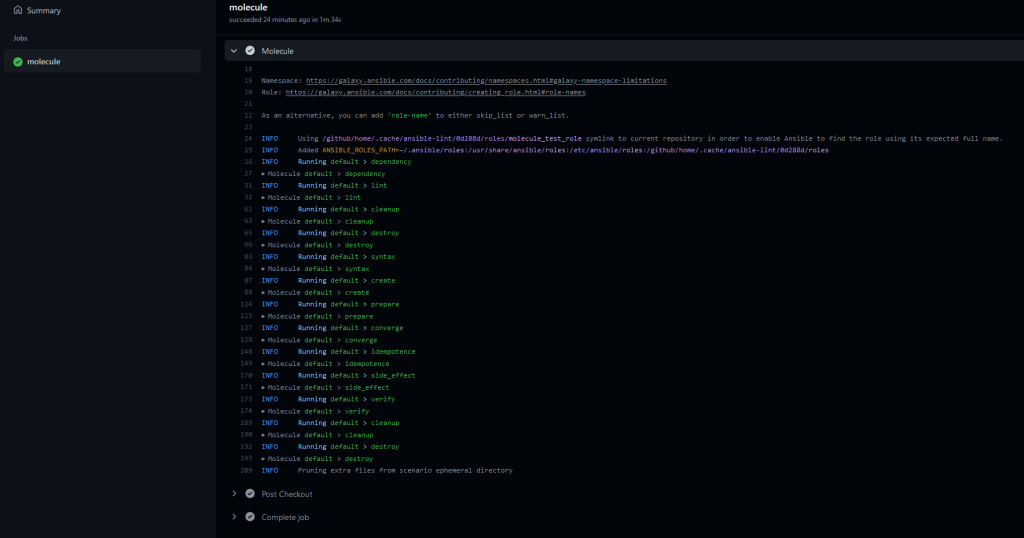
And this is really it. By now you should have a fully automated Ansible Molecule test running, every time you push to your repository.
Philip
UPDATE 24.04.2022: I’ve decided for me, that I don’t rely on the molecule action anymore. They introduced some breaking changes for me and I wasn’t able to fix them in a reasonable amount of time. So I decided to build my own action setup. You can find the latest version also in my Github repository. In the end, my molecule.yml looks now like this:
---
name: Molecule
on:
push:
branches:
- master
pull_request:
branches:
- master
jobs:
molecule:
runs-on: ubuntu-latest
steps:
- name: Checkout
uses: actions/checkout@v2
- name: Set up Python
uses: actions/setup-python@v2
with:
python-version: '3.x'
- name: Install dependencies.
run: pip3 install yamllint ansible-lint ansible "molecule[lint,docker]"
- name: Install Galaxy dependencies.
run: ansible-galaxy collection install community.docker
- name: Run molecule
run: "cd ansible/molecule/molecule_test_role && molecule test"
As you can see, I setup a basic Ubuntu system and install all dependencies on my own. Also I run the molecule test now by commandline instead of calling the Github action.





Hi, I’m a maintainer of github action gofrolist/molecule-action.
Thank you for using my product and write a good article about it.
Just curious what issue you had with molecule-action?
Hi Gofrolist,
thanks for stepping by. The issue I had with the action was that the ansible-lint command could not be found.
/bin/sh: ansible-lint: not foundI haven’t found time to troubleshoot this really, my action manifest looked like this:
name: Moleculeon:
push:
branches:
- master
pull_request:
branches:
- master
jobs:
molecule:
runs-on: ubuntu-latest
steps:
- name: Checkout
uses: actions/checkout@v2
with:
path: "${{ github.repository }}"
- name: Molecule
uses: gofrolist/molecule-action@v2.2.30
with:
molecule_command: test
molecule_args: -d docker
molecule_working_dir: thedatabaseme/cookbooks/ansible/molecule/molecule_test_role
env:
ANSIBLE_FORCE_COLOR: '1'
Hope to hear you soon.
Philip
I’ve got issue request to add `ansible-list` and it has been added recently by this commit https://github.com/gofrolist/molecule-action/pull/111/files#diff-230078d672f10d17463a8a6265cad825b790885898256a3365be90685caac58dR7
For now it’s in master and it will be released soon.
Thanks for the information. I will watch the repo and test it out. Thanks for your good work.